Research Project
A Mutational Study of Residues Critical for Protein Stability in the Fingers and Palm Subdomains of HIV-1 Reverse Transcriptase
Advisor: Dr. Clyde A. Hutchison III, University of North Carolina
Abstract:
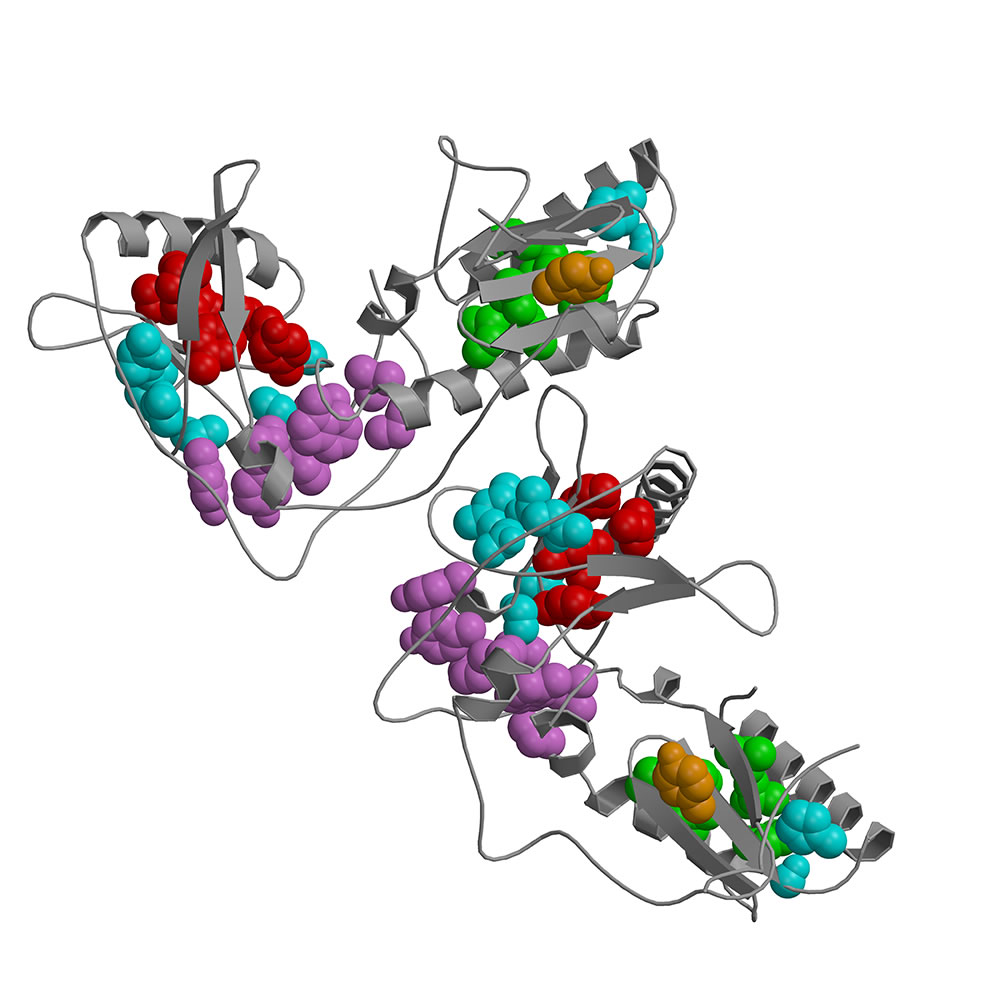
This study uses a mutational approach to identify residues critical for protein stability in a large heterodimeric protein with multiple subdomains, the HIV-1 reverse transcriptase. We constructed 526 single amino acid replacement mutations in the fingers and palm subdomains of the HIV-1 reverse transcriptase. Screening each mutant using two simple assays (a polymerase activity assay and Western blot analysis) allowed us to identify mutations that destabilized the protein. With our large data set, we were able to experimentally explore the interactions of residues critical for protein stability in a large protein. We found both hydrophobic and hydrophilic residues critical for protein stability. The critical hydrophobic residues tend to cluster in the interior of the protein, buried in a non-polar environment. These critical hydrophobic residues grouped into three cores, one in fingers subdomain, one in the palm subdomain, and one in the junction between the fingers and palm subdomains. We found that there is a higher degree of mutational specificity for hydrophobic residues in these hydrophobic cores, compared to the other hydrophobic residues in the protein. We also identified hydrophilic residues involved in H-bonds that appear to play a role in protein stability. These critical hydrophilic residues are segregated from, but near, the hydrophobic cores. A number of these H-bonds appear to form a connection between the hydrophobic core in the fingers subdomain and the hydrophobic core in the fingers-palm junction. These H-bonds may also have a critical role in protein stability by bringing together secondary structural elements.
Residues critical for protein stability in HIV-1 Reverse Transcriptase
(fingers and palm subdomains)
PUBLICATIONS:
Gunawardena, H.P., Feltcher, M.E., Wrobel, J.A., Gu, S., Braunstein, M., and Chen, X. (2013). Comparison of the Membrane Proteome of Virulent Mycobacterium tuberculosis and the Attenuated Mycobacterium bovis BCG Vaccine Strain by Label-Free Quantitative Proteomics. Journal of Proteome Research 12, 5463-5474. [ PubMed ]
Khatun, J., Yu, Y., Wrobel, J.A., Risk, B.A., Gunawardena, H.P., Secrest, A., Spitzer, W.J., Ling, X., Wang, L., Chen, X., and Giddings, M.C. (2013). Whole human genome proteogenomic mapping for ENCODE cell line data: identifying protein-coding regions. BMC Genomics 14, 141. [ PubMed ]
The ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. [ PubMed ]
Djebali, S. et al. (2012). Landscape of transcription in human cells. Nature 489, 101-108. [ PubMed ]
Wrobel, J.A., Conrad, M.J., Bloedon, E., Swanstrom, R., and Hutchison, C.A., III. (2000). Analysis of HIV type 1 Reverse Transcriptase: Comparing Sequences of Viral Isolates with Mutational Data. AIDS Research and Human Retroviruses 16, 2049-2054. [ PubMed ] [ Supplemental material ]
Wrobel, J.A., Chao, S.-F, Conrad, M.J., Merker, J.D., Swanstrom, R., Pielak, G.J., and Hutchison, C.A., III. (1998). A Genetic Approach for Identifying Critical Residues in the Fingers and Palm Subdomains of HIV-1 Reverse Transcriptase. Proc. Natl. Acad Sci. USA 95, 638-645. [ PubMed ]
Wrobel, J.A. (1996). Saturation Mutation and Crystal Structure Analysis of the HIV-1 Reverse Transcriptase Fingers and Palm Subdomains. Ph.D. dissertation, University of North Carolina, Chapel Hill.
Hoffmann, G.R., Sprague, K.M., Wrobel, J.A., and Wroblewski, D.H. (1987). A Recombinagenic Effect of Strychnine in Salmonella typhimurium. Environmental and Molecular Mutagenesis 10, 27-33. [ PubMed ]